Guinea Pig anti-Rabbit IgG (Heavy & Light Chain) Antibody - Preadsorbed
-
- Key Features
-
- 95+ publication references, 1 independent validation
- Frequently used as CUT&RUN IgG negative control and CUT&Tag secondary antibody
- Used in CUT&RUN and CUT&Tag protocols, e.g. Henikoff et al. (2018) PMID 29651053, Kaya-Okur et al. (2019, 2020) PMID 31036827 and PMID 32913232
- Target See all IgG products
- IgG
-
Binding Specificity
- Heavy & Light Chain
-
Reactivity
- Rabbit
-
Host
- Guinea Pig
-
Clonality
- Polyclonal
-
Application
- ELISA, Immunohistochemistry (IHC), Western Blotting (WB), Cleavage Under Targets and Tagmentation (CUT&Tag), Cleavage Under Targets and Release Using Nuclease (CUT&RUN)
- Purpose
-
The Guinea Pig anti-Rabbit IgG antibody ABIN101961 is well suited as a CUT&RUN IgG negative control and as a secondary antibody in CUT&Tag. It is a component of all our CUT&RUN Product Sets.
Find more products for CUT&RUN and CUT&Tag:
- Specificity
- Rabbit IgG (H&L)
- Cross-Reactivity (Details)
- No reaction was observed against Human, Mouse and Goat Serum Proteins.
- Purification
- Preadsorption: Solid phase absorption
- Sterility
- Sterile filtered
- Immunogen
- whole molecule of rabbit IgG
- Isotype
- IgG
-
-
- Application Notes
-
The guinea pig anti-rabbit IgG antibody ABIN101961 is suitable for use in ELISA, immunohistochemistry, and Western Blot, CUT&RUN and CUT&Tag. Specific conditions for each assay should be optimized by the end user. General ABIN101961 dilution recommendations for different applications are as follows:
- ELISA: 1:20,000 - 1:100,000
- WB: 1:2,000 - 1:10,000
- IHC: 1:1,000 - 1:5,000
- CUT&RUN: 1:100
- CUT&Tag: 1:100
- Comment
-
ABIN101961 is tested via ELISA to ensure that the titer against the antigen (Rb IgG) is above a certain threshold. We also test to make sure the titer against potentially cross-reactive human IgG, goat IgG, and mouse IgG is below a certain threshold.
In addition, we test ABIN101961 against anti-guinea pig Serum, rabbit IgG, and rabbit serum in an immunoelectrophoresis assay. - Restrictions
- For Research Use only
-
- by
- Tom Taghon’s lab, Vakgroep Diagnostische Wetenschappen, Universiteit Gent
- No.
- #104174
- Date
- 12/18/2019
- Antigen
- rabbit IgG
- Lot Number
- 43586
- Method validated
- Cleavage Under Targets and Tagmentation
- Positive Control
rabbit anti-H3K27me3 monoclonal antibody
- Negative Control
rabbit normal IgG antibody
- Notes
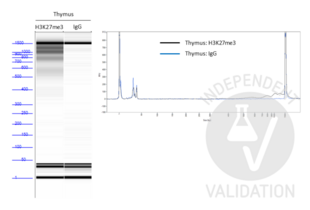
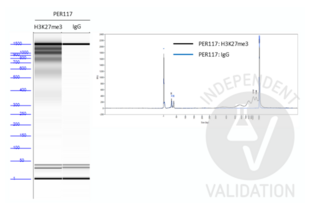
Passed. ABIN101961 successfully increased the number of protein A binding sites in a CUT&Tag protocol on human primary thymocytes and PER-117 cells using a monoclonal rabbit H3K27me3 primary antibody.
- Primary Antibody
- rabbit IgG anti-H3K27me3 antibody (Cell Signaling Technology, 9733T, lot 14)
- Secondary Antibody
- ABIN101961
- Full Protocol
- Profiling of H3K27me3 signals in human primary CD34+ sorted thymocytes and PER-117 cell line (50,000 cells each).
- Carry out the CUT&Tag protocol according to the single-tube bench top protocol for CUT&Tag developed by Steven Henikoff’s lab as outlined on the protocols.io platform.
- Use reagents as suggested in the original protocol. The pA-Tn5 fusion protein used in this validation was a courtesy of Steven Henikoff’s lab and used at 1:1000 dilution for 100 µL per sample.
- Primary antibody binding with
- 1µl/sample monoclonal rabbit IgG anti-H3K27me3 antibody (Cell Signaling Technology, 9733T, lot 14) or
- 1µl/sample rabbit normal IgG (Cell Signaling, 2729S, lot 9).
- Secondary antibody binding with 1µl/sample guinea pig anti rabbit antibody (antibodies-online, ABIN101961, lot 43586).
- Determine library concentration on a Qubit Fluorometer using Qubit dsDNA High Sensitivity Assay Kit (ThermoFisher Scientific, Q32581).
- Determine library size distribution of all samples on an Agilent Fragment Analyzer using High Sensitivity Small DNA Fragment Analysis Kit (Agilent, DNF-477-0500).
- Experimental Notes
ABIN101961 successfully increased the number of protein A binding sites for each bound rabbit anti-H3K27me3 antibody in the human primary thymocytes and PER-117 cell line. This resulted in quantifiable amounts of tagmented genomic fragments after PCR amplification that showed a ladder-like distribution.
- Library concentrations as measured on a Qubit Fluorometer were 0.122ng/µl and 0.538ng/µl for thymocytes and PER-117 cells respectively using a rabbit H3K27me3 primary antibody. Library concentration for both sample was below the instrument’s detections limit using the rabbit normal IgG primary antibody.
- As discussed at the protocols.io, it is expected to observe a ladder-like distribution for H3K27me3 profiling. However, using lower cell numbers could alter the nucleosomal patterns and result in an increase in larger fragments. This is observed by Steven Henikoff’s lab as well. Nevertheless, it does not affect the quality of the sequencing results.
Validation #104174 (Cleavage Under Targets and Tagmentation)![Successfully validated 'Independent Validation' Badge]()
![Successfully validated 'Independent Validation' Badge]() Validation ImagesFull Methods
Validation ImagesFull Methods -
- by
- Cantù Lab, Gene Regulation during Development and Disease, Linköping University
- No.
- #104622
- Date
- 02/10/2024
- Antigen
- IgG
- Lot Number
- 48661
- Method validated
- Cleavage Under Targets and Release Using Nuclease
- Positive Control
Anti H3K4me (antibodies-online, ABIN3023251)
- Negative Control
Guinea Pig anti-rabbit IgG (antibodies-online, ABIN101961)
- Notes
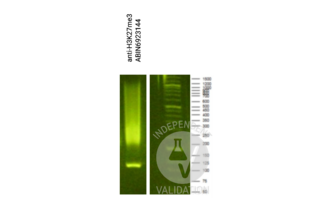
Passed.
- Primary Antibody
- ABIN101961
- Secondary Antibody
- Full Protocol
- Cell harvest and nuclear extraction
- Harvest 250,000 HEK293T cells per antibody
- Centrifuge cell solution 5 min at 600 x g at RT.
- Remove the liquid carefully.
- Gently resuspend cells in 2 mL of Nuclear Extraction Buffer (20 mM HEPES-KOH pH 8.2, 20% Glycerol, 0.05% IGEPAL, 0.5 mM Spermidine, 10 mM KCl, Roche Complete Protease Inhibitor EDTA-free).
- Move the solution to a 2 mL centrifuge tube.
- Pellet the nuclei 800 x g for 5 min.
- Repeat the NE wash twice for a total of three washes.
- Resuspend the nuclei in 40 µL NE Buffer per sample.
- Concanavalin A beads preparation
- Prepare one 2 mL microcentrifuge tube.
- Gently resuspend the magnetic Concanavalin A Beads (antibodies-online, ABIN6923139).
- Pipette 10 µL Con A Beads slurry for each sample into the 2 mL microcentrifuge tube.
- Place the tube on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Pipette 1 mL Binding Buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2) into the tube and resuspend ConA beads by gentle pipetting.
- Spin down the liquid from the lid with a quick pulse in a table-top centrifuge.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Repeat the wash twice for a total of three washes.
- Gently resuspend the ConA Beads in 44 µL Binding Buffer.
- Nuclei immobilization – binding to Concanavalin A beads
- Carefully vortex the nuclei suspension and add 44 µL of the Con A beads in Binding Buffer to the cell suspension for each sample.
- Close tube tightly and incubate 10 min at 4 °C.
- Put the 2 mL tube on the magnet stand and when the liquid is clear, remove the supernatant.
- Resuspend the beads in 1 mL of EDTA wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, Roche Complete Protease Inhibitor EDTA-free, 2 mM EDTA).
- Incubate 5 min at RT.
- Resuspend the beads in 200 µL of wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, Roche Complete Protease Inhibitor EDTA-free).
- Primary antibody binding
- Divide nuclei suspension into separate 200 µL PCR tubes, one for each antibody.
- Add 2 µL antibody (anti-H3K4me positive control ABIN3023251, and guinea pig anti-rabbit IgG negative control antibody ABIN101961) to the respective tube, corresponding to a 1:100 dilution.
- Incubate at 4 °C overnight.
- Wash with 200 µL of Wash Buffer using a multichannel pipette to accelerate the process.
- Repeat the wash five times for a total of six washes.
- pAG-MNase Binding
- Prepare a 1.5 mL microcentrifuge tube containing 200 µL of pAG mix per sample (200 µL of wash buffer + 120 ng pAG-MNase per sample).
- Place the PCR tubes with the sample on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Resuspend the beads in 200 µL of pAG-MNase premix.
- Incubate 30 min at 4 °C.
- Wash with 200 µL of Wash Buffer using a multichannel pipette to accelerate the process.
- Repeat the wash five times for a total of six washes.
- Resuspend in 100 µL of Wash Buffer.
- MNase digestion and release of pAG-MNase-antibody-chromatin complexes
- Place PCR tubes on ice and allow to chill.
- Prepare a 1.5 mL microcentrifuge tube with 102 µL of 2 mM CaCl2 mix per sample (100 µL Wash Buffer + 2 µL 100 mM CaCl2) and let it chill on ice.
- Resuspend the sample in 50 µL of 1x Urea STOP Buffer (8.5 M Urea, 100 mM NaCl, 2 mM EGTA, 2 mM EDTA, 0.5% IGEPAL).
- Incubate the samples 1h at 4 °C.
- Transfer the supernatant containing the pAG-MNase-bound digested chromatin fragments to fresh 200 µL PCR tubes.
- DNA Clean up (Mag-Bind® TotalPure NGS - M1378-01)
- Add 2x volume of beads to each sample (e.g. 100 µL of beads for 50 µL of sample).
- Wash with 80% EtOH.
- Resuspend the beads in 20 µL of 10 mM Tris.
- Library preparation and sequencing
- Prepare Libraries using KAPA HyperPrep Kit using KAPA Dual-Indexed adapters according to protocol.
- Sequence samples on an Illumina NextSeq 500 sequencer, using a NextSeq 500/550 High Output Kit v2.5 (75 Cycles), 36bp PE.
- Peak calling
- Trim reads using bbTools bbduk to remove adapters, artifacts, and repeat sequences.
- Aligned reads were mapped to the hg38 human genome using bowtie with options -m 1 -v 0 -I 0 -X 500.
- Use SAMtools to convert SAM files to BAM files and remove duplicates.
- Use BEDtools genomecov to produce Bedgraph files.
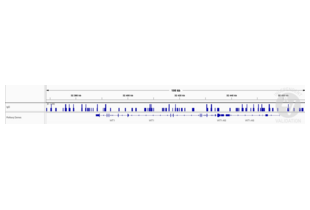
- Call peaks using SEACR with a 0.001 threshold and the option norm stringent.
- Cell harvest and nuclear extraction
- Experimental Notes
Validation #104622 (Cleavage Under Targets and Release Using Nuclease)![Successfully validated 'Independent Validation' Badge]()
![Successfully validated 'Independent Validation' Badge]() Validation ImagesFull Methods
Validation ImagesFull Methods -
- Format
- Liquid
- Concentration
- 1.02 mg/mL
- Buffer
- 0.02 M Potassium Phosphate, 0.15 M Sodium Chloride, pH 7.2, 0.01 % (w/v) NaN3, no stabilizer
- Preservative
- Sodium azide
- Precaution of Use
- This product contains Sodium azide: a POISONOUS AND HAZARDOUS SUBSTANCE which should be handled by trained staff only.
- Storage
- 4 °C,-20 °C
- Storage Comment
- Store vial at 4° C prior to opening. This product is stable for several weeks at 4° C as an undiluted liquid. Dilute only prior to immediate use. For extended storage aliquot contents and freeze at -20° C or below.
- Expiry Date
- 12 months
-
-
: "Chem-map profiles drug binding to chromatin in cells." in: Nature biotechnology, (2023) (PubMed).
: "The ETS transcription factor ETV6 constrains the transcriptional activity of EWS-FLI to promote Ewing sarcoma." in: Nature cell biology, Vol. 25, Issue 2, pp. 285-297, (2023) (PubMed).
: "Histone remodeling reflects conserved mechanisms of bovine and human preimplantation development." in: EMBO reports, Vol. 24, Issue 3, pp. e55726, (2023) (PubMed).
: "The Hdc GC box is critical for Hdc gene transcription and histamine-mediated anaphylaxis." in: The Journal of allergy and clinical immunology, (2023) (PubMed).
: "Loss of MLL3/4 decouples enhancer H3K4 monomethylation, H3K27 acetylation, and gene activation during embryonic stem cell differentiation." in: Genome biology, Vol. 24, Issue 1, pp. 41, (2023) (PubMed).
: "An integrative epigenomic approach identifies ELF3 as an oncogenic regulator in ASCL1-positive neuroendocrine carcinoma" in: Cancer Science, (2023) (PubMed).
: "Transcriptional-translational conflict is a barrier to cellular transformation and cancer progression." in: Cancer cell, (2023) (PubMed).
: "Heat shock transcription factors demonstrate a distinct mode of interaction with mitotic chromosomes." in: Nucleic acids research, (2023) (PubMed).
: "BIND&MODIFY: a long-range method for single-molecule mapping of chromatin modifications in eukaryotes." in: Genome biology, Vol. 24, Issue 1, pp. 61, (2023) (PubMed).
: "Combinatory EHMT and PARP inhibition induces an interferon response and a CD8 T cell-dependent tumor regression in PARP inhibitor-resistant models." in: bioRxiv : the preprint server for biology, (2023) (PubMed).
: "Engineered MED12 mutations drive uterine fibroid-like transcriptional and metabolic programs by altering the 3D genome compartmentalization." in: Research square, (2023) (PubMed).
: "RNA Polymerase II, the BAF remodeler and transcription factors synergize to evict nucleosomes." in: bioRxiv : the preprint server for biology, (2023) (PubMed).
: "Analyzing the Genome-Wide Distribution of Histone Marks by CUT&Tag in Drosophila Embryos." in: Methods in molecular biology (Clifton, N.J.), Vol. 2655, pp. 1-17, (2023) (PubMed).
: "DNA selection by the master transcription factor PU.1." in: Cell reports, Vol. 42, Issue 7, pp. 112671, (2023) (PubMed).
: "Cancer lineage-specific regulation of YAP responsive elements revealed through large-scale functional epigenomic screens." in: Nature communications, Vol. 14, Issue 1, pp. 3907, (2023) (PubMed).
: "Engineered MED12 mutations drive leiomyoma-like transcriptional and metabolic programs by altering the 3D genome compartmentalization." in: Nature communications, Vol. 14, Issue 1, pp. 4057, (2023) (PubMed).
: "Tet2 regulates Sin3a recruitment at active enhancers in embryonic stem cells." in: iScience, Vol. 26, Issue 7, pp. 107170, (2023) (PubMed).
: "Context-defined cancer co-dependency mapping identifies a functional interplay between PRC2 and MLL-MEN1 complex in lymphoma." in: Nature communications, Vol. 14, Issue 1, pp. 4259, (2023) (PubMed).
: "H3K36 methylation maintains cell identity by regulating opposing lineage programmes." in: Nature cell biology, Vol. 25, Issue 8, pp. 1121-1134, (2023) (PubMed).
: "PLA2G2E-mediated lipid metabolism triggers brain-autonomous neural repair after ischemic stroke." in: Neuron, (2023) (PubMed).
-
: "Chem-map profiles drug binding to chromatin in cells." in: Nature biotechnology, (2023) (PubMed).
-
- Target
- IgG
- Abstract
- IgG Products
- Target Type
- Antibody
-

 (105 references)
(105 references) (2 validations)
(2 validations)




