Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the enveloped positive-sense RNA viruses. This virus is characterized by club-like spikes on the surface, and a unique replication strategy. Cell entry of coronaviruses depends on binding of the viral spike (S) proteins to cellular receptors and on S protein priming by host cell proteases. Unravelling which cellular factors are used by SARS-CoV-2 for entry might provide insights into viral transmission and reveal therapeutic targets.1
In the following the replication cycle of SARS-CoV-2 is explained together with possible inhibitors and their respective targets. This compilation is based on current literature however we make no claim to accuracy.
Stages of the SARS-CoV-2 Life Cycle:
SARS-CoV-2 Replication Cycle
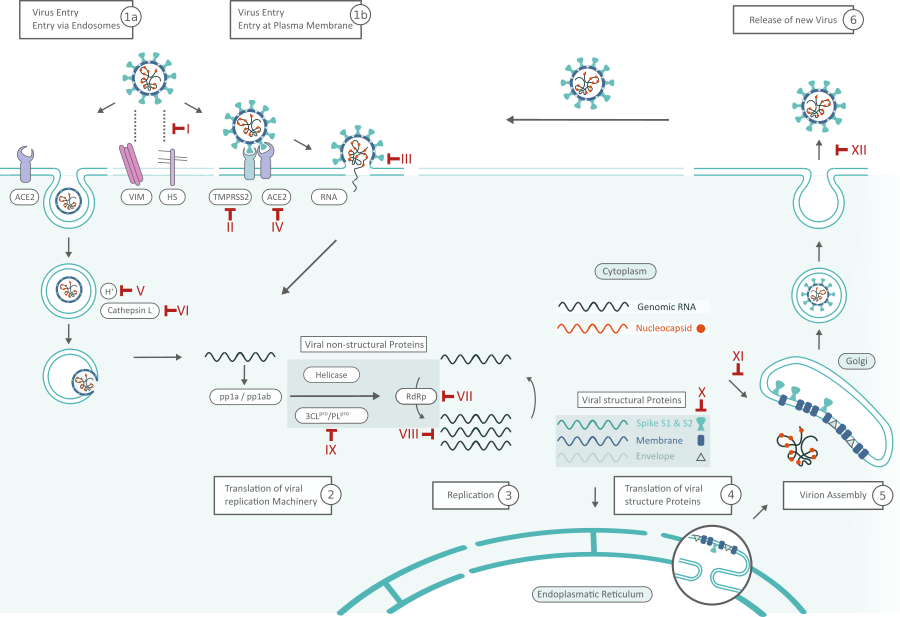
SARS-Cov-2 Replication Cycle and Inhibitors. Possible targets for inhibitors are marked in red and numbered in roman numerals. Click here for full PDF version
Virus Entry (1)
SARS-CoV-2 can hijack the cell in two ways, either via endosomes or via plasma membrane fusion. (In both ways) Spike proteins (S1, S2) of SARS-CoV-2 mediate attachment to the membrane of a host cell and engage angiotensin-converting enzyme 2 (ACE2) as the entry receptor.1 Inhibitors like Griffithsin (Inhibitor III) bind to the spike glycoprotein, thus preventing viral entry. Cell surface vimentin (VIM) acts as a critical co-receptor and is essential for successful ACE-2 binding.2 Binding of heparan sulfate (HS) to the receptor binding domain (RBD) enhances binding to ACE2 as well. Viral adhesion may be inhibited by by exogenous heparin. Heparin competes with HS for binding of the SARS-CoV-2 S protein.3
When virions are taken up into endosomes, cathepsin L activates the spike protein. The pH dependent cysteine protease can be blocked by lysosomotropic agents, like bafilomycin A1 or ammonium chloride (Inhibitor Classes IV,V). Alternatively, the spike protein can be cleaved between the S1 and S2 domains by the cellular serine protease TMPRSS2 in close proximity to the ACE2 receptor, which initiates fusion of the viral membrane with the plasma membrane (Inhibitor II: Camostat). 1 The plasma membrane fusion entry is less likely to trigger host cell antiviral immunity and therefore more efficient for viral replication. 4
SARS-CoV-2 Inhibitor Screening Kit
This inhibitor screening ELISA pair is designed to facilitate the identification and characterization of SARS-CoV-2 inhibitors.
Show product detailsRelated Products: SARS-CoV-2 Spike Proteins | SARS-CoV-2 Neutralizing Antibodies | SARS-CoV-2 S1 Protein Mutations | ACE2 Antibodies | TMPRSS2 Antibodies
- (4)
- (5)
- (1)
- (1)
- (2)
- (3)
- (1)
Translation of Viral Replication Machinery (2) and Replication (3)
After the viral RNA is released into the host cell, polyproteins are translated. The coronavirus genomic RNA encodes nonstructural proteins (NSPs) that have a critical role in viral RNA synthesis, and structural proteins which are important for virion assembly. First, polyproteins pp1a and pp1ab, are translated which are cleaved by the Papain-like protease (PLpro, Nsp3) and 3C-like protease (3CLpro, Nsp5) (Inhibitor VIII) to form functional NSPs such as Helicase or the RNA replicase–transcriptase complex (RdRp).5 RdRp especially can be inhibited by virostatica like Favipiravir or Penciclovir (Inhibitor VI); the replication of viral RNA in general by kinase signaling pathway inhibitors like Saracatinib (Inhibitor VII). The expression level of N protein can be decreased by resveratrol (Inhibitor X).7
One of the first translated proteins is the host shutoff factor Nsp1. This viral protein interferes with translation and causes accelerated degradation of host mRNA, thus suppressing the host’s innate immune response. 8
Related Products: SARS-CoV-2 Non-Structural Proteins | SARS-CoV-2 N Proteins | SARS-CoV-2 N Antibodies
SARS-CoV-2 S Mutation Proteins
We support your research with reliable SARS-CoV-2 mutation proteins. B.1.1.7 / P.1 / B.1.351 / S Protein wild type. 400+ labs in diagnostics, pharma, academia have used our SARS-CoV-2 research tools.
Explore our variety of mutation proteins- (9)
- (8)
- (1)
- (2)
- (3)
- (2)
Translation of Viral Structure Proteins (4) and Virion Assembly (5)
RdRp (Nsp12) is responsible for replication of structural protein RNA. Structural proteins S, Envelope (E), Membrane (M) are translated by ribosomes that are bound to the endoplasmic reticulum (ER). The ER forms double membrane vesicles (DMVs) in which the viral RNA is replicated and shielded from the host’s innate immune system. Nsp3 creates pores through which viral RNA leaves the DMVs for virion assembly. The nucleocapsid proteins (N) remain in the cytoplasm and are assembled from genomic RNA. They fuse with the virion precursor which is then transported from the ER through the Golgi Apparatus to the cell surface via small vesicles.
Related Products: SARS-CoV-2 Spike Proteins | SARS-CoV-2 S1 Protein Mutations | SARS-CoV-2 S Antibodies
- (8)
- (15)
- (1)
- (5)
- (3)
Release of Virus (6)
Virions are then released from the infected cell through exocytosis and search another host cell. Oseltamivir inhibits cleavage of sialic acids by neuroamidase from the cell receptors thus preventing release of newly formed virions from the cell surface (Inhibitor XI). One feature that sets SARS-CoV-2 apart from other coranaviruses like e.g. SARS-CoV-2 is a second cleavage site in the S protein. Furin cleavage at the S1/S2 site probably takes place when virions are released through the Golgi apparatus or lysosomes. It primes the S protein for a second cut at the S2’ site by TMPRSS2. Certain mutations within this site are also hallmarks of SARS-CoV-2 variants of concern alpha (B.1.1.7), beta (B.1.351), delta (B.1.617.2) and omicron (B.1.1.529). 8
Related Products:
Need Help? Call our PhD Customer Support!
- We help you with finding the right product for your research.
- We offer reliable antibodies, kits, proteins, lysates for COVID-19 research.
- Contact us via email or phone: (877) 302 8632 (US) or +49 241 95 163 153 (International)
Related Information and Products
SARS-CoV-2 Neutralizing Antibodies based on CR3022
Mutations of SARS-CoV-2 S Protein
SARS-CoV Protein Interactome / poster and products
SARS-CoV-2 RT-PCR Kits and qPCR Kits
References
- (1) Hoffmann et al.: SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell (2020), PDF
- Suprewicz et al.: Vimentin binds to SARS-CoV-2 spike protein and antibodies targeting extracellular vimentin block in vitro uptake of SARS-CoV-2 virus-like particles, BioRxiv preprint (2021), [DOI]
- (2) Henderson et al: Controlling the SARS-CoV-2 Spike Glycoprotein Conformation, Nat Struct Mol Biol. (2021), [ DOI]
- (3) Shirato, K., Kawase, M. & Matsuyama, S.: Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry, Virology 517, 9–15 (2018), [DOI]
- (4) Zhavoronkov et al.: Potential COVID-2019 3C-like Protease Inhibitors Designed Using Generative Deep Learning Approaches, ChemRxiv (2020), [DOI]
- (5) Shin et al.: Saracatinib Inhibits Middle East Respiratory Syndrome-Coronavirus Replication In Vitro, Viruses, 10(6):283 (2018), [DOI]
- (6) Lin, S. C., Ho, C. T., Chuo, W. H., Li, S., Wang, T. T., & Lin, C. C. : Effective inhibition of MERS-CoV infection by resveratrol, BMC Infectious Diseases, 17(1)(2017), [DOI]
- (7) McKimm‐Breschkin: Influenza neuraminidase inhibitors: Antiviral action and mechanisms of resistance. Influenza and Other Respiratory Viruses 7(Suppl. 1), 25–36., [PMC]
- (8) Scudellari M.: How the coronavirus infects cells — and why Delta is so dangerous, Nature (2021), [DOI]

Creative mind of antibodies-online with a keen eye for details. Proficient in the field of life-science with a passion for plant biotechnology and clinical study design. Responsible for illustrated and written content at antibodies-online as well as supervision of the antibodies-online scholarship program.
Go to author page



