Amyotrophic Lateral Sclerosis
Amyotrophic lateral sclerosis (ALS) is a late-onset fatal motor neuron disease (MND) with an incidence of about 1-5/100,000 that was first described in 1869 by the French neurologist Jean-Martin Charcot (Charcot disease) and became well known in the late 1930 when baseball player Lou Gehrig was diagnosed with it (Lou Gehrig’s disease).
Most ALS cases are sporadic (SALS), but 5–10% of the cases are familial ALS (FALS) with an underlying genetic cause. Typically, symptoms emerge around 60 years of age for SALS and around 50 years for FALS. Both SALS and FALS caused by a progressive degeneration of cortical and spinal motor neurons, leading to paralysis and ultimately respiratory failure because of muscle impairment. Mutations in numerous genes have been associated with mechanisms causing neuronal degeneration such as RNA dysregulation, impaired protein homeostasis, or the formation of cytoplasmic aggregates. Many of these genetic marks are shared with the closely related disease frontotemporal dementia (FTD). Non-neuronal cells, such as astrocytes and microglia, exacerbate neurodegeneration via the secretion of neurotoxic mediators and the modulation of glutamate receptor expression. Dysfunction of oligodendrocytes and Schwann cells cause damage to the myelin.
Besides genetic predisposition, environmental factors are thought to contribute to developing ALS given that the disease typically manifests in adulthood. Suspected risk factors among others include smoking, head trauma, athletic propensity, and neurotoxic chemicals, e.g. b-N-methylamino-L-alanine (BMMA).
So far, there is no effective treatment for ALS. The principal options are neuroprotective treatment, symptomatic and supportive treatment as well as some unproven disease-modifying therapies such as stem cell transplantation and gene therapy.
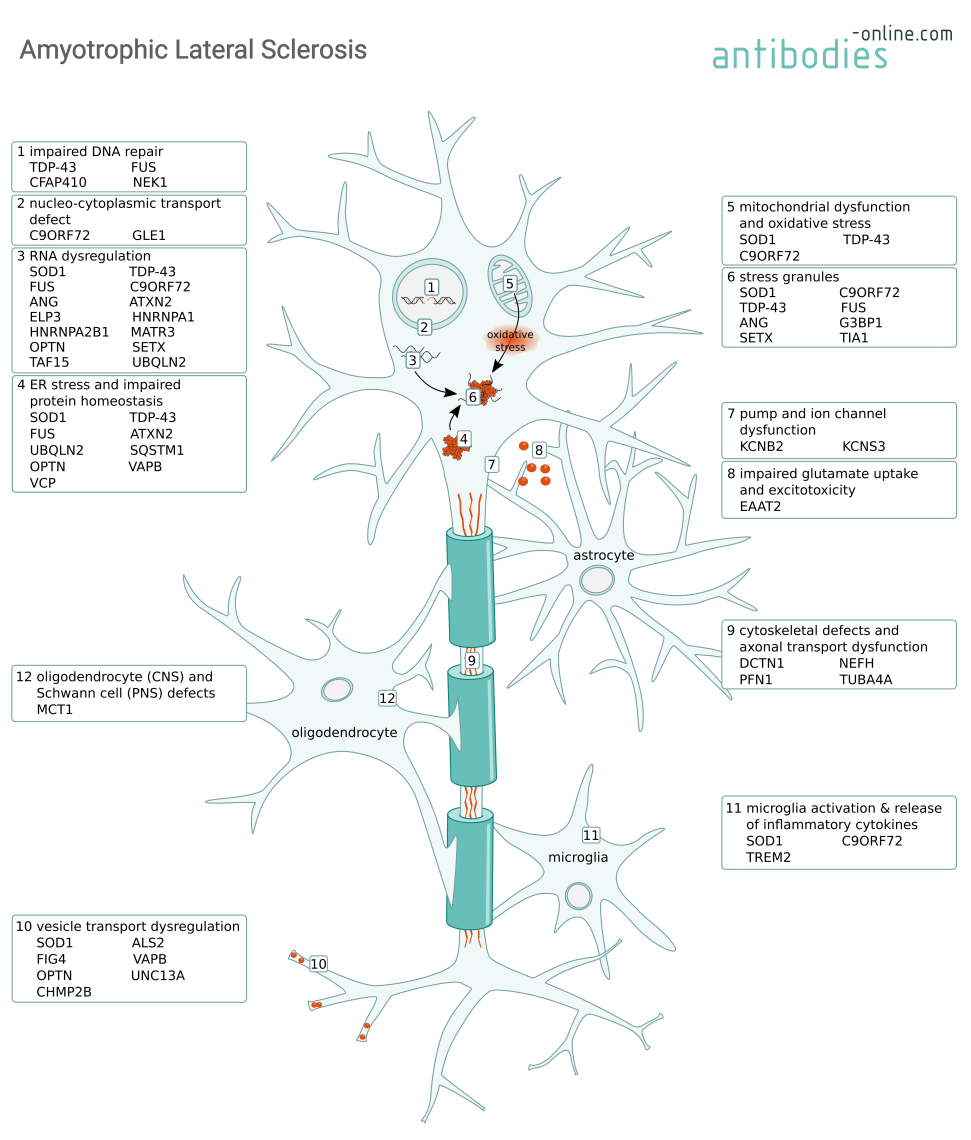
Mechanisms causing motor neuron degeneration in ALS and some of the involved proteins.
Important Targets Related to Amyotrophic Lateral Sclerosis
More than 140 genes have been identified to cause, to be associated with ALS, or to influence disease progression, including:
C9ORF72
C9ORF72 is a protein abundant in neurons that part of the C9ORF7-SMCR protein complex. It is thought to have guanine nucleotide exchange factor activity, regulate autophagy, and to play a role in endosomal trafficking. Mutations in the C9orf72 gene are most frequently associated with ALS (up to 50% of FALS and 10% of SALS cases).
Three mechanisms are thought to contribute to the ALS pathology depending on C9ORF72 mutations. Firstly, the expansion of a GGGGCC repeat within the C9orf72 promotor leads to decreased protein expression and consequently a loss of function. Secondly, the accumulation of GGGGCC repeat-containing RNA transcripts leads to the formation of toxic RNA aggregates which further sequester additional RNA-binding proteins and lead to dysregulation of the protein homeostasis and the formation of stress granules. And thirdly, a toxic gain of function when the GGGGCC hexanucleotide repeats are translated into dipeptide repeat (DPR) containing proteins. These poly-GA, poly-GP, poly-GR, poly-PA, and poly-PR peptides (depending on the reading frame) form DPR inclusions that impair nucleocytoplasmic transport and can cause neurodegeneration. These proposed mechanisms are not mutually exclusive an their relative contributions are still being investigated.
C9ORF72 Antibodies
SOD1
Genetic studies have identified mutations in superoxide dismutase (SOD1) as the second-most common cause for FALS with a proportion of 10-20%.
SOD1 is a highly expressed primarily cytosolic enzyme that catalyzes the conversion of superoxide into hydrogen peroxide and oxygen. Mutant SOD1 binds to mitochondria and compromises respiration by blocking protein import. This causes oxidative stress, an imbalance in the cellular Ca2+ homeostasis, the formation of intracellular aggregates and stress granules. These aggregates lead to neurofilament aggregation and adversely affect axonal transport processes. Ultimately, mutant SOD1 can induce apoptosis via Bcl-2 inhibition.
SOD1 Antibodies
TDP-43
Mutations in the TAR-DNA binding protein 43 kDa (TDP-43), encoded by the TARDBP gene, are less common than the C9ORF72 DPR or the SOD1 mutations. However, its mislocalization to the cytoplasm is a characteristic feature in many ALS cases.
TDP-43 is an essential DNA- and RNA-binding nuclear protein involved in regulating transcription, modulating gene splicing, RNA metabolism, and stress granules. In both familial and sporadic ALS and other neurodegenerative diseases such as the closely related Frontotemporal Dementia (FTD) or Alzheimer's Disease (AD), mutated TDP-43 forms are a major component of the insoluble ubiquitinated protein aggregates in the cytoplasm. These TDP-43 aggregates sequester miRNAs and proteins, thus impairing protein homeostasis. Dysregulation of nuclear-encoded mitochondrial proteins leads to mitochondrial dysfunction and oxidative stress.
Knocking down TDP-43 is not a viable therapeutic approach because of the proteins’ critical cellular function. It has however been shown that decreasing ataxin 2 (ATXN2) expression using antisense oligonucleotides (ASOs) also reduces TDP-43 toxicity.
TDP-43/TARDBP Antibodies
FUS
Similar to TDP-43, fused in sarcoma (FUS) is an RNA-binding protein involved in transcription an RNA processing. Consistent with its function, wild type FUS is located in the nucleus. In ALS however, mutated forms of the protein are mislocalized to the cytoplasm. Here, gain of function mutations can cause aggregation of FUS and associated RNA binding proteins, thus leading to an imbalance in protein homeostasis, RNA dysregulation, and the formation of stress granules which ultimately lead to motor neuron loss.
FUS mutations are present in approximately 5% of FALS and less than 2% of SALS cases. They are frequently associated with early-onset ALS. Because of the dose-dependent toxic effect of mutated FUS variants the protein is a potential therapeutic target for antisense oligonucleotide silencing.
FUS Antibodies
References
- : "Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS." in: Nature, Vol. 466, Issue 7310, pp. 1069-75, (2010) (PubMed).
- : "A comprehensive review of amyotrophic lateral sclerosis." in: Surgical neurology international, Vol. 6, pp. 171, (2015) (PubMed).
- : "An amyotrophic lateral sclerosis-linked mutation in GLE1 alters the cellular pool of human Gle1 functional isoforms." in: Advances in biological regulation, Vol. 62, pp. 25-36, (2017) (PubMed).
- : "Decoding ALS: from genes to mechanism." in: Nature, Vol. 539, Issue 7628, pp. 197-206, (2016) (PubMed).
- : "Pathology of Neurodegenerative Diseases." in: Cold Spring Harbor perspectives in biology, Vol. 9, Issue 7, (2018) (PubMed).
- : "The Role of Dipeptide Repeats in C9ORF72-Related ALS-FTD." in: Frontiers in molecular neuroscience, Vol. 10, pp. 35, (2017) (PubMed).
- : "Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice." in: Nature, Vol. 544, Issue 7650, pp. 367-371, (2017) (PubMed).
- : "Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder." in: The EMBO journal, Vol. 36, Issue 20, pp. 2931-2950, (2017) (PubMed).
- : "ALS Genes in the Genomic Era and their Implications for FTD." in: Trends in genetics : TIG, Vol. 34, Issue 6, pp. 404-423, (2019) (PubMed).
- : "Amyotrophic Lateral Sclerosis: An Update for 2018." in: Mayo Clinic proceedings, Vol. 93, Issue 11, pp. 1617-1628, (2019) (PubMed).
- : "RNA Dysregulation in Amyotrophic Lateral Sclerosis." in: Frontiers in genetics, Vol. 9, pp. 712, (2018) (PubMed).
- : "Impaired NHEJ repair in amyotrophic lateral sclerosis is associated with TDP-43 mutations." in: Molecular neurodegeneration, Vol. 15, Issue 1, pp. 51, (2021) (PubMed).
- : "Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis." in: Molecular cell, Vol. 80, Issue 5, pp. 876-891.e6, (2020) (PubMed).
- : "Stress Granule Dysregulation in Amyotrophic Lateral Sclerosis." in: Frontiers in cellular neuroscience, Vol. 14, pp. 598517, (2020) (PubMed).
- : "Improving clinical trial outcomes in amyotrophic lateral sclerosis." in: Nature reviews. Neurology, Vol. 17, Issue 2, pp. 104-118, (2021) (PubMed).
- : "TDP-43 aggregation induced by oxidative stress causes global mitochondrial imbalance in ALS." in: Nature structural & molecular biology, Vol. 28, Issue 2, pp. 132-142, (2021) (PubMed).
- : "Oligodendrocyte Dysfunction in Amyotrophic Lateral Sclerosis: Mechanisms and Therapeutic Perspectives." in: Cells, Vol. 10, Issue 3, (2021) (PubMed).
- : "Microglial TREM2 in amyotrophic lateral sclerosis." in: Developmental neurobiology, Vol. 82, Issue 1, pp. 125-137, (2022) (PubMed).
- : "TREM2 interacts with TDP-43 and mediates microglial neuroprotection against TDP-43-related neurodegeneration." in: Nature neuroscience, Vol. 25, Issue 1, pp. 26-38, (2022) (PubMed).
- : "Antisense oligonucleotide silencing of FUS expression as a therapeutic approach in amyotrophic lateral sclerosis." in: Nature medicine, Vol. 28, Issue 1, pp. 104-116, (2022) (PubMed).
- : "Apolipoprotein A1 Enhances Endothelial Cell Survival in an In Vitro Model of ALS." in: eNeuro, Vol. 9, Issue 4, (2022) (PubMed).
- : "Elevated α5 integrin expression on myeloid cells in motor areas in amyotrophic lateral sclerosis is a therapeutic target." in: Proceedings of the National Academy of Sciences of the United States of America, Vol. 120, Issue 32, pp. e2306731120, (2023) (PubMed).

Goal-oriented, time line driven scientist, proficiently trained in different academic institutions in Germany, France and the USA. Experienced in the life sciences e-commerce environment with a focus on product development and customer relation management.
Go to author page



