Mutations of SARS-CoV-2 S Protein
Genomic mutations play a key role in propagation of SARS-COV-2 and virus in general. They may facilitate infection and pose an additional challenge for the detection by the host cell. This turns them into important research targets especially in the context of vaccine and drug design.
- Incl. N501Y Mutation.
- Source: HEK-293 Cells.
- For ELISA, Ligand Binding Assay.
- In Stock, Fast Delivery.
- Incl. N501Y, E484K.
- Source: HEK-293 Cells
- For ELISA, Ligand Binding Assay.
- In Stock, Fast Delivery.
- T19R, L452R, T478K, D614G, P681R, D950N
- For ELISA, Ligand Binding Assay.
- Source: HEK-293 Cells
- In Stock, Fast Delivery.
- Incl. N501Y, Q498R, H655Y, N679K and P681H.
- Source: HEK-293 Cells
- For ELISA, Ligand Binding Assay.
- In Stock, Fast Delivery.
- Wild type, full length Cov-2 spike protein.
- For assay development.
- In Stock, Fast Delivery.
Mutations in B.1.1.7 and B.1.351 / 501Y.V2 Lineage
Mutation N501Y is a non-synonymous mutation within the S-protein’s receptor binding domain (RBD) shared by the two SARS-CoV-2 lineages B.1.1.7 and B.1.351 first identified in southeastern England and South Africa respectively. It is one of the key contact residues within the RBD and has been identified as increasing binding affinity to human and murine ACE2. Currently there is no evidence suggesting that the mutations in these variants have any impact on the severity of disease or vaccine efficacy. It has however been speculated that they lead to higher concentrations in the upper airways and thus increase the virus’ transmission rate by around 50%.
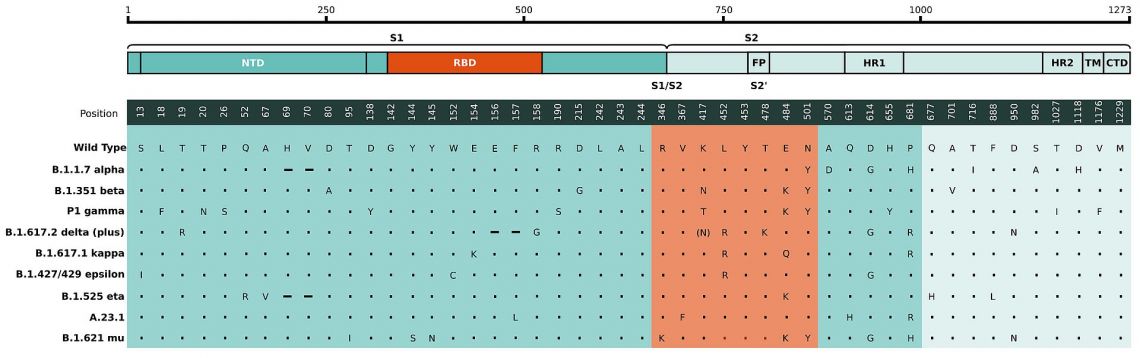
Fig. 1: a) Mutations in the viral spike identified in B.1.351 / 501Y.V2 (SA) and B.1.1.7 (UK) in addition to D614G.11 b) Amino acid changes in the spike region of the 190 S501Y.V2 genomes mapped to the spike protein sequences structure, indicating key regions, such as the RBD. Each spike protein variant is shown at their respective protein locations, with the bar lengths representing the number of genomes harboring the specific mutations (Only mutations that appear in >10% of sequences are shown). The D614G mutation is already present in the parent lineage.9
The E484K mutation, present in the novel lineages 501Y.S2 and B.1.1.28 from South Africa and Brazil respectively, affects a residue within the RBD that has been shown to be important for binding of many neutralizing antibodies. Accordingly, this mutation affects antibody recognition and enable SARS-CoV-2 immune escape. Virus bearing this mutation has been shown to escape recognition by antibodies in peoples’ convalescent sera and may thus alter the effectiveness of vaccines. 7,8,9
D614G Mutation
The mutation D614G (Asp614Gly) has become dominant in the european region indicating a fitness advantage relative to the original Wuhan strain that enables more rapid spread. Several recent studies indicate that the D614G mutation diminishes the interaction between the S1 and S2 units, facilitating the shedding of S1 from viral-membrane-bound S2. This leads to an increase of total S protein incorporated into the virion and therefore more stable virus particles.1,2,3 We also offer SARS-CoV-2 S protein D614G mutant in a trimeric form, facilitating researchers for more detailed virological and immunological studies on the biological effect of the mutation.
- (4)
- (4)
- (4)
Other Mutations of the Spike Protein
Several other mutations have been discovered. The mutations V367F (Val367Phe), N354D (Asn354Asp), W436R (Trp436Arg) or V483A (Val483Ala) of the S1 protein have been shown to bind with higher affinity to ACE2.4,5 V483A and G476S (Gly476Ser) mutations have previously been reported to be related to human receptor-binding affinity in MERS and SARS-CoV research.10 R408I (Arg408Ile) on the other hand potentially reduce the ACE2 binding affinity.6
- (1)
- (3)
- (1)
- (3)
- (4)
- (1)
- (3)
Protein Structure of some Mutation S-Proteins
Fig.2 SARS-CoV-2 S Protein B.1.1.7 (alpha)
Fig.3 SARS-CoV-2 S Protein B.1.351 (beta)
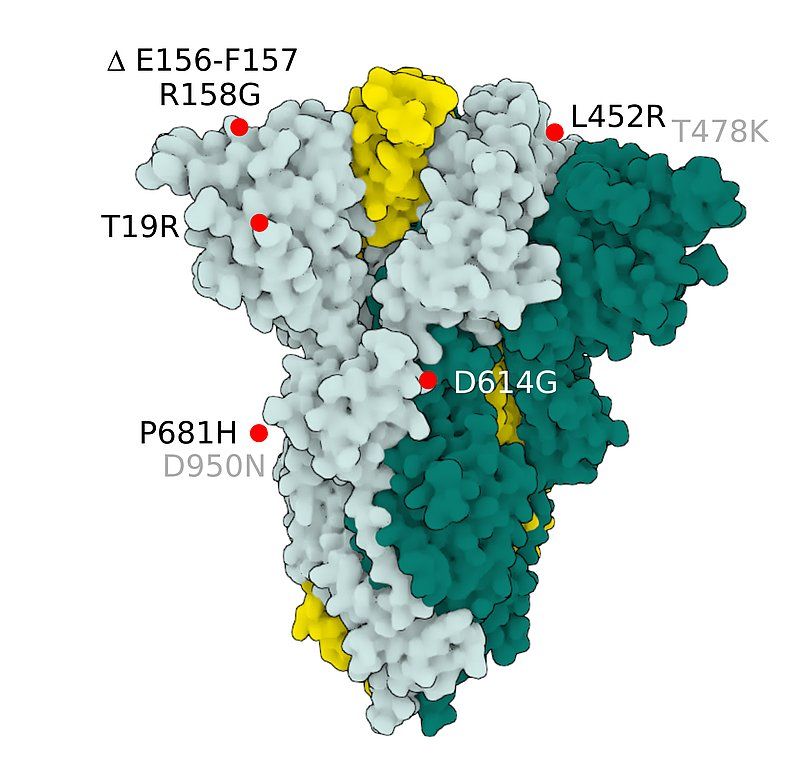
Fig.4 SARS-CoV-2 S Protein B.1.617.2 (delta)
Fig.5 SARS-CoV-2 S Protein P.1 (gamma)
References
- (1) Jie Hu et al. The D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity and decreases neutralization sensitivity to individual convalescent sera. bioRxviv (2020).
- (2) Korber B. et al.Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxviv (2020). doi.org/10.1101/2020.04.29.069054.
- (3) Lizhou Zhang et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxviv (2020). doi.org/10.1101/2020.06.12.148726.
- (4) Junxian Ou et al. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein. bioRxiv (2020). doi:10.1101/2020.03.15.991844v4
- (5) Saha, P. et al.Mutations in Spike Protein of SARS-CoV-2 Modulate Receptor Binding, Membrane Fusion and Immunogenicity: An Insight into Viral Tropism and Pathogenesis of COVID-19. chemRxiv (2020). doi:10.26434/chemrxiv.12320567.v1
- (6) Jian Shang, Yushun Wan, Chuming Luo, Gang Ye, Qibin Geng, Ashley Auerbach, Fang Li. Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences May 2020, 117 (21) 11727-11734; DOI: 10.1073/pnas.2003138117
- (7) Allison J. Greaney, Andrea N. Loes, Katharine H.D. Crawford, Tyler N. Starr, Keara D. Malone, Helen Y. Chu, Jesse D. Bloom, bioRxiv 2020.12.31.425021; doi: https://doi.org/10.1101/2020.12.31.425021
- (8) Nicholas G. Davies, Rosanna C. Barnard, Christopher I. Jarvis, Adam J. Kucharski, James Munday, Carl A. B. Pearson, Timothy W. Russell, Damien C. Tully, Sam Abbott, Amy Gimma, William Waites, Kerry LM Wong, Kevin van Zandvoort, CMMID COVID-19 Working Group, Rosalind M. Eggo, Sebastian Funk, Mark Jit, Katherine E. Atkins, W. John Edmunds. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv 2020.12.24.20248822; doi: https://doi.org/10.1101/2020.12.24.20248822
- (9) Houriiyah Tegally, Eduan Wilkinson, Marta Giovanetti, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.12.21.20248640; doi: https://doi.org/10.1101/2020.12.21.20248640
- (10) Kim JS, Jang JH, Kim JM, Chung YS, Yoo CK, Han MG. Genome-Wide Identification and Characterization of Point Mutations in the SARS-CoV-2 Genome. Osong Public Health Res Perspect. 2020;11(3):101-111. doi:10.24171/j.phrp.2020.11.3.05
- (11) Pengfei Wang, Lihong Liu, Sho Iketani, Yang Luo et al. Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv preprint posted January 26, 2021. ; https://doi.org/10.1101/2021.01.25.428137doi.

Goal-oriented, time line driven scientist, proficiently trained in different academic institutions in Germany, France and the USA. Experienced in the life sciences e-commerce environment with a focus on product development and customer relation management.
Go to author page



