SARS-CoV-2 Proteins
antibodies-online provides a large selection of recombinant proteins for SARS-CoV-2 research and assay development including membrane protein, nucleocapsid protein, spike protein, S protein mutations, envelope protein, and non-structural proteins. Discover our product portfolio.
Top Products: Trimeric SARS-CoV-2 Spike protein
- (2)
- (6)
- (5)
- (4)
- (4)
- (4)
*Super Stable Trimer: Proline substitutions (F817P, A892P, A899P, A942P, K986P, V987P) and alanine substitutions (R683A and R685A) are introduced to stabilize the trimeric prefusion state of SARS-CoV-2 S protein and abolish the furin cleavage site, respectively.
SARS-CoV-2 Spike (S1, S2) Protein
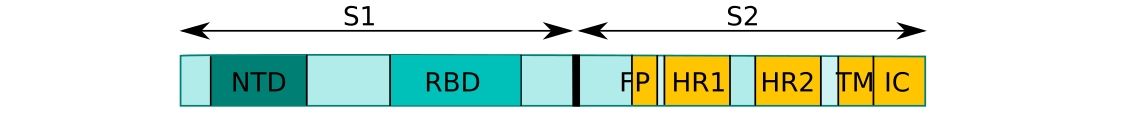
CoV uses its spike glycoprotein (S), a main target for neutralization antibody, to bind its receptor, and mediate membrane fusion and virus entry. Each monomer of trimeric S protein is about 180 kDa, and contains two subunits, S1 and S2, mediating attachment and membrane fusion, respectively.6 Below you can find a schematic drawing of SARS-CoV-2 Spike protein: NTD, N-terminal domain. FP, fusion prptide. HR1, heptad repeat 1. HR2, heptad repeat 2. TM, transmembrane domain.16
- (9)
- (6)
- (4)
- (5)
- (4)
- (3)
- (3)
- (3)
- (3)
- (2)
- (2)
- (2)
- (2)
Click here to see all SARS-CoV-2 Spike Protein Products
SARS-CoV-2 Nucleocapsid (N) Protein
The nucleocapsid protein is an important structural protein for the coronaviruses. It is highly abundant in the viruses. Its function involves entering the host cell, binding to the viral RNA genome, and forms the ribonucleoprotein core.(7) N protein contains two distinct RNA-binding domains (NTD and CTD) linked by a poorly structured linkage region containing a serine/arginine-rich (SR-rich) domain.
- (5)
- (2)
- (1)
- (2)
- (1)
- (1)
- (1)
- (1)
- (1)
- (2)
- (2)
- (2)
SARS-CoV-2 Membrane (M) Protein
The coronavirus membrane (M) protein is the key player in virion assembly. One of its functions is to mediate the incorporation of the spikes into the viral envelope. When expressed alone, it accumulates in the Golgi complex in homomultimeric complexes.
SARS-CoV-2 Envelope (E) Protein
E protein of SARS-CoV-2 is a 75 amino acids long protein existing in both monomeric and homo-pentameric form. Approximately 20 copies of the protein have been found in the viral particle and previous mutagenesis-based studies demonstrated its pivotal role in the onset and development of the viral infection.9
- (1)
- (2)
- (1)
SARS-CoV-2 Non-structural Proteins (NSP)
The SARS-CoV-2 genome encodes 16 non-structural proteins (Nsp1-16), four structural proteins, and nine putative accessory factors.(8) NSPs include the various enzymes and transcription factors the virus uses to replicate itself, such as viral protease, RNA replicase and proteins to control the host.
- (1)
- (1)
- (1)
- (1)
- (1)
- (1)
- (1)
- (1)
- (1)
The role of recombinant Proteins in SARS-CoV-2 Research
Recombinant SARS-CoV-2 proteins are being used as antigens for antibody development, as capture antigens or as standards in assays. They can be used as a positive control in antigen-detecting ELISAs to accurately separate true positive results from potentially false results or as capture antigens for immunoglobin ELISAs. The trimeric, full length SARS-CoV-2 Spike protein for example is suitable for assay development and highly useful when studying neutralizing antibodies. In addition SARS-CoV-2 proteins are needed for drug discovery and drug repurposing studies. In the process of drug discovery, functional studies with active proteins are vital to verify the inhibitory effects of the tested substance. NSPs as well as N proteins are in the spotlight as potential targets; their functions and interaction with the host cell are crucial for virus propagation and therefore highly relevant for inhibition strategies. SARS-CoV-2 N protein has been shown to affect the complement system whereas the SARS-CoV-2 NSPSs are responsible for virus replication.
Related Products: full length SARS-CoV-2 Spike protein (ABIN6952670)
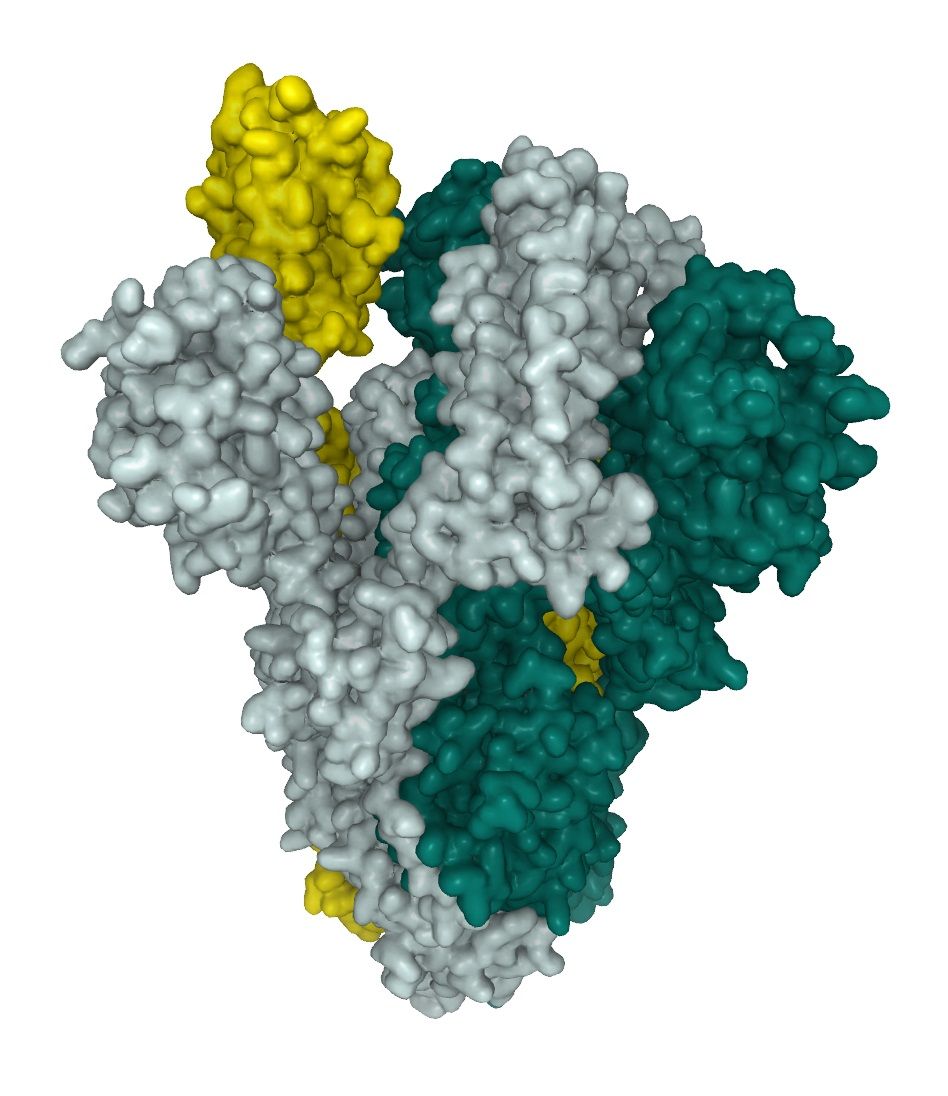
Post-translational modifications (PTMs), like glyscosilation, modify proteins as last step of maturation to promote protein folding and improve stability. The glycosylation pattern of the SARS-CoV-2 spike protein provides basic understanding of the viral structure, and is important for the identification of immunogens for vaccine design, especially regarding steric hindrance. The spike glycoprotein exists as a homotrimeric fusion protein. Each of the trimers contains 66 glycosylation sites for host-derived N-linked glycans. In the predominant state of the trimer, one of the RBDs is in an “up” position whereas the other two are in a “down” position. Interaction of S-protein and ACE2 only takes place with one RBD in the “up” position.
SARS-CoV-2 utilizes high mannose as well as complex-type glycans structure on their spike proteins (see below). 1 This leads to a complex surface structures, a challenge for finding interactors and in the generation of neutralizing antibodies (Nabs). The Full length SARS-CoV-2 Spike protein (ABIN6952670) is, like our other active SARS-CoV-2 proteins produced in HEK293-cells. The mammalian expression system is able to mimic complex glycosilation pattern of SARS-CoV-2 and express the homotrimeric fusion protein in its active "up" state (fig.1). The Full length SARS-CoV-2 Spike protein therefore is highly useful when studying neutralizing antibodies.
In a recent Glycobiology article Shajahan et al. performed site-specific quantitative N-linked and O-linked glycan profiling on recombinant SARS-CoV-2 S protein subunit S1 and SARS-CoV-2 S protein subunit S2 through glycoproteomics using high resolution LC-MS/MS. The spike protein is comprised of two protein subunits (S1 and S2), which together possess 22 potential N-glycosylation sites. The group identified 2 unexpected O-glycosylation sites at the receptor binding domain (RBD) of subunit S1.1
Related Products: SARS-CoV-2 Spike Subunit S1 (AA 16-690) protein (His tag) (ABIN6952319) | SARS-CoV-2 Spike Subunit S2 (AA 697-1213) protein (His tag) (ABIN6952319)
The N-glycans on S protein play important roles in proper protein folding and priming by host proteases. Since glycans can shield the amino acid residues and other epitopes from cells and antibody recognition, glycosylation can enable the coronavirus to evade both the innate and adaptive immune responses.2,3,4 The group used recombinant SARS-CoV-2 S1 Protein and SARS-CoV-2 S2 Protein expressed in HEK293 cells and observed partial N-glycan occupancy on 17 out of 22 N-glycosylation sites. High mannose-type (Man5GlcNAc2) sugar chains were implemented as predominant structure across all sites.5
Thr323 and Ser325 were identified as O-glycosylation sites on the S1 subunit of SARS-CoV-2 spike protein through high resolution mass spectrometry glycoproteomic profiling. The residues Thr323 and Ser325 are located at the RBD of the S1 subunit of SARS-CoV-2, and thus the O-glycosylation at this location could play a critical role in viral binding with hACE2 receptors.3
References
: "Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2." in: Glycobiology, Vol. 30, Issue 12, pp. 981-988, (2020) (PubMed).: "Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology." in: Cell, Vol. 183, Issue 4, pp. 1024-1042.e21, (2020) (PubMed).
: "The proximal origin of SARS-CoV-2." in: Nature medicine, Vol. 26, Issue 4, pp. 450-452, (2020) (PubMed).
: "Global aspects of viral glycosylation." in: Glycobiology, Vol. 28, Issue 7, pp. 443-467, (2018) (PubMed).
: "Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV." in: Nature communications, Vol. 11, Issue 1, pp. 1620, (2020) (PubMed).
: "Biochemical characterization of SARS-CoV-2 nucleocapsid protein." in: Biochemical and biophysical research communications, Vol. 527, Issue 3, pp. 618-623, (2020) (PubMed).
: "A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing." in: bioRxiv : the preprint server for biology, (2020) (PubMed).
: "Immunoinformatic analysis of the SARS-CoV-2 envelope protein as a strategy to assess cross-protection against COVID-19." in: Microbes and infection, Vol. 22, Issue 4-5, pp. 182-187, (2020) (PubMed).
: "Cell entry mechanisms of SARS-CoV-2." in: Proceedings of the National Academy of Sciences of the United States of America, Vol. 117, Issue 21, pp. 11727-11734, (2020) (PubMed).
- Jie Hu et al. The D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity and decreases neutralization sensitivity to individual convalescent sera. bioRxviv (2020).
- Korber B. et al.Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxviv (2020). doi.org/10.1101/2020.04.29.069054.
- Lizhou Zhang et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxviv (2020). doi.org/10.1101/2020.06.12.148726.
- Junxian Ou et al. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein. bioRxiv (2020). doi:10.1101/2020.03.15.991844v4
- Saha, P. et al.Mutations in Spike Protein of SARS-CoV-2 Modulate Receptor Binding, Membrane Fusion and Immunogenicity: An Insight into Viral Tropism and Pathogenesis of COVID-19. chemRxiv (2020). doi:10.26434/chemrxiv.12320567.v1

Creative mind of antibodies-online with a keen eye for details. Proficient in the field of life-science with a passion for plant biotechnology and clinical study design. Responsible for illustrated and written content at antibodies-online as well as supervision of the antibodies-online scholarship program.
Go to author page



