SARS-CoV-2 Interferon Antagonism
Recent studies on SARS-CoV-2 evaluating evasion of immune response reveal several SARS-CoV-2 proteins which manipulate host response in favor of virus proliferation. Compared with other virus-related respiratory diseases, the interferon I (IFN-I) and interferon III (IFN-III) signaling is more efficiently suppressed.1 This ultimatively leads to low interferon-stimulated gene (ISG) responses with impact on viral transmission and pathogenesis.3
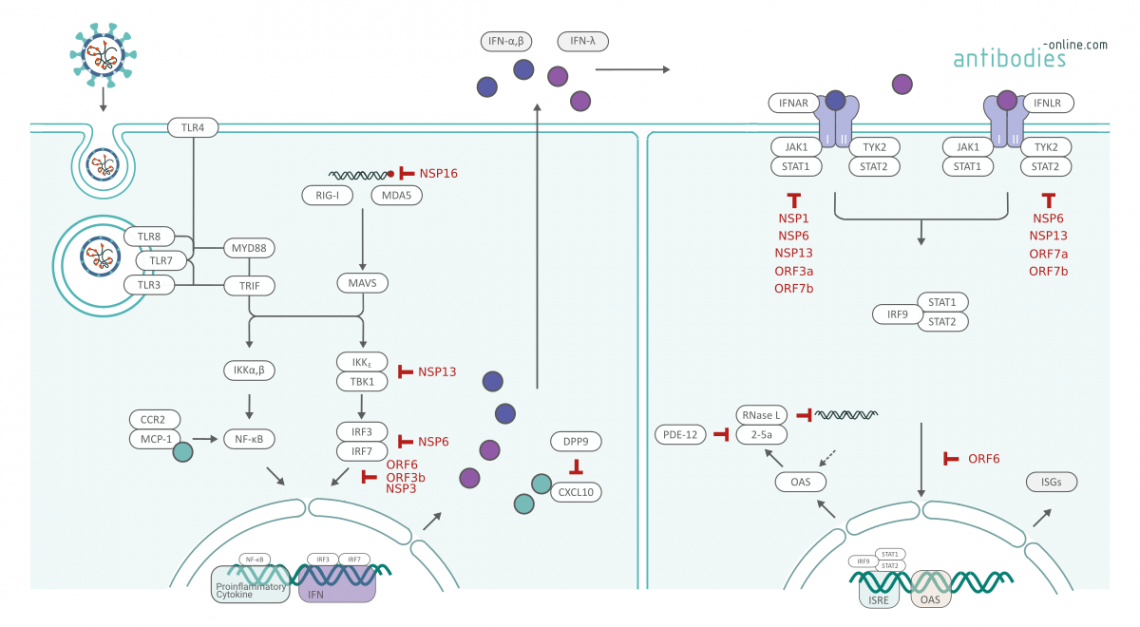
Interferon Antagonism of SARS-Cov-2 Proteins according to Xia et al. and Park et al.2
Interferon Antagonism related Antibodies
- (1)
- (8)
- (2)
- (4)
- (2)
- (3)
- (1)
- (6)
- (6)
- (1)
- (5)
- (1)
- (2)
- (3)
- (1)
SARS-CoV-2 antagonize IFN-I Production and Signaling
An unbiased screening of SARS-CoV-2 proteins identified main antagonist to IFN-I response. Several SARS-CoV-2 proteins antagonize IFN-I production via distinct mechanisms: Similar to SARS-CoV, NSP16 of SARS-CoV-2 is able to modify the 5′ cap with its 2’-O-methyl-transferase activity, allowing the virus to efficiently evade recognition by melanoma differentiation-associated protein 5 (MDA5). 1,6 NSP13 binds and blocks TANK binding kinase 1 (TBK1) phosphorylation, Nonstructural protein 6 (NSP6) binds TBK1 to suppress interferon regulatory factor 3 (IRF3) phosphorylation and ORF6 binds importing Karyopherin α 2 (KPNA2) to inhibit IRF3 nuclear translocation.2 IRF3 nuclear translocation is further inhibited by ORF3b and NSP3, the truncated ORF3b of SARS-CoV-2 suppresses IFN induction more efficiently than that of SARS-CoV, which may contribute to the poor IFN response reported in COVID-19 patients.1,4
Two sets of viral proteins antagonize IFN-I and IFN-III (IL28a/IL28b/IL29) signaling through blocking of signal transducer and activator of transcription 1 (STAT1)/STAT2 phosphorylation or nuclear translocation. NSP1, ORF3a and ORF7b antagonize STAT1 signaling, ORF7a and ORF7b antagonizes STAT2 signaling.2 NSP6, and NSP13 are key actors in interferon antagonism. They suppress both signaling pathways alongside with IRF3 nuclear translocation. Taken together, SARS-CoV-2 inhibits IFN-I signaling through suppression and inhibition of STAT1 and STAT2 phosphorylation and through blockign of STAT1 nuclear translocation. This ultimatively leads to low expression of ISGs and a limited antiviral response.
Interferon Antagonism related Antibodies
- (2)
- (7)
- (3)
- (6)
- (16)
- (8)
- (3)
- (1)
- (2)
- (2)
- (1)
- (2)
- (3)
- (2)
- (4)
- (1)
Potential therapeutic SARS-CoV-2 Targets
Pairo-Castineira et al used Mendelian randomisation to find potential targets for repurposing of licensed medications. A low expression of Interferon-alpha/beta receptor beta chain (IFNAR2), and high expression of TYK2 and C-C chemokine receptor type 2 (CCR2) is associated with critical illness. TYK2 is a target for JAK inhibitors already in medical use. The receptor CCR2 is able to bind monocyte chemotactic protein 1 (MCP-1). MCP-1 concentrations are associated with more severe disease. Anti-CCR2 monoclonal antibody therapy in treatment of rheumatoid arthritis is safe. 6
Dipeptidyl peptidase 9 (DPP9) encodes a serine protease with diverse intracellular functions, including cleavage of the key antiviral signalling mediator CXCL10 and key roles in antigen presentation, and inflammasome activation.
Another potential target is therapeutic target is PDE-12. Upon contact with viral dsDNA, OAS1 produces 2'-5'A which activates an effector enzyme, RNAse L. RNAse L degrades double-stranded RNA which consequently activates MDA5 leading to interferon production. Endogenous or exogenus phosphodiesterase 12 (PDE-12) activity degrades the host antiviral mediator 2-5A. 6,7
Interferon Antagonism related SARS-CoV-2 Proteins
- (1)
Related Resources
References
- : "Type I and Type III Interferons - Induction, Signaling, Evasion, and Application to Combat COVID-19." in: Cell host & microbe, Vol. 27, Issue 6, pp. 870-878, (2020) (PubMed).
- : "SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses." in: Cell, Vol. 183, Issue 5, pp. 1325-1339.e21, (2020) (PubMed).
- : "Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19." in: Cell, Vol. 181, Issue 5, pp. 1036-1045.e9, (2020) (PubMed).
- : "Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV." in: Journal of virology, Vol. 94, Issue 23, (2020) (PubMed).
- : "SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant." in: Cell reports, Vol. 32, Issue 12, pp. 108185, (2020) (PubMed).
- : "Genetic mechanisms of critical illness in COVID-19." in: Nature, Vol. 591, Issue 7848, pp. 92-98, (2021) (PubMed).
- : "The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors." in: The Journal of biological chemistry, Vol. 290, Issue 32, pp. 19681-96, (2015) (PubMed).

Goal-oriented, time line driven scientist, proficiently trained in different academic institutions in Germany, France and the USA. Experienced in the life sciences e-commerce environment with a focus on product development and customer relation management.
Go to author page



