Histone 3 antibody (H3K4me)
-
- Target See all Histone 3 (H3) Antibodies
- Histone 3 (H3)
-
Binding Specificity
- H3K4me
-
Reactivity
- Human
-
Host
- Rabbit
-
Clonality
- Polyclonal
-
Conjugate
- This Histone 3 antibody is un-conjugated
-
Application
- Western Blotting (WB), Immunofluorescence (IF), Chromatin Immunoprecipitation (ChIP), Immunoprecipitation (IP), ChIP DNA-Sequencing (ChIP-seq), Cleavage Under Targets and Release Using Nuclease (CUT&RUN)
- Cross-Reactivity
- Human, Mouse, Rat
- Characteristics
- Methylated Antibodies
- Purification
- Affinity purification
- Immunogen
- A synthetic methylated peptide corresponding to residues surrounding K4 of human histone H3
- Isotype
- IgG
-
-
- Application Notes
- WB 1:500 - 1:2000, IF 1:50 - 1:200, IP 1:50 - 1:200, ChIP 1:50 - 1:200, ChIP-seq 1:50 - 1:200, CUT&RUN 1:100
- Restrictions
- For Research Use only
-
- by
- Mattias Pernebrink, Anna Nordin and Claudio Cantù; Cantù Lab, Gene Regulation during Development and Disease, Linköping University
- No.
- #104228
- Date
- 11/12/2021
- Antigen
- H3K4me
- Lot Number
- 3560036504
- Method validated
- Cleavage Under Targets and Release Using Nuclease
- Positive Control
- Recombinant anti-H3K27me3 CUT&RUN Positive Control antibody (antibodies-online, ABIN6923144)
- Negative Control
- Guinea Pig anti-rabbit IgG (antibodies-online, ABIN101961)
- Notes
Passed. ABIN3023251 allows for H3K4me targeted digestion using CUT&RUN.
- Primary Antibody
- ABIN3023251
- Secondary Antibody
- Full Protocol
- Cell harvest
- Harvest 250,000 HEK293T cells per antibody to be used at RT.
- Centrifuge cell solution 3 min at 600 x g at RT.
- Remove the liquid carefully.
- Gently resuspend cells in 1 mL Wash Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, Roche Complete Protease Inhibitor EDTA-free) by pipetting and transfer cell solution to a 2 mL microcentrifuge tube.
- Centrifuge cell solution 3 min at 600 x g at RT and discard the supernatant.
- Repeat twice for a total of three washes.
- Resuspend cell pellet in 1 mL Wash Buffer by gently pipetting.
- Concanavalin A beads preparation
- Prepare one 1.5 mL microcentrifuge tube.
- Gently resuspend the magnetic Concanavalin A Beads (antibodies-online, ABIN6923139).
- Pipette 10 µL Con A Beads slurry for each sample into the 1.5 mL microcentrifuge tube.
- Place the tube on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Pipette 1 mL Binding Buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2) into each tube and resuspend ConA beads by gentle pipetting.
- Spin down the liquid from the lid with a quick pulse in a table-top centrifuge.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Repeat twice for a total of three washes.
- Gently resuspend the ConA Beads in a volume of Binding Buffer corresponding to the original volume of bead slurry, i.e. 10 µL per sample.
- Cell immobilization – binding to Concanavalin A beads
- Carefully vortex the cell suspension and add 10 µL of the Con A beads in Binding Buffer to the cell suspension for each sample.
- Close tube tightly and rotate for 10 min at RT.
- Cell permeabilization and primary antibody binding
- Divide cell suspension into separate 2 mL microcentrifuge tubes, one for each antibody (250,000 cells per sample).
- Place the microcentrifuge tubes on a magnetic stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Place each tube at a low angle on the vortex mixer set to a low speed and add 150 µL Digitonin Wash buffer (wash buffer with 0.025% (wt/vol) Digitonin) supplemented with 2 mM EDTA.
- Gently vortex the microcentrifuge tubes until the beads are resuspended.
- o Add 1.5 µL antibody (anti-H3K4me ABIN3023251, anti-H3K27me3 positive control antibody ABIN6923144, or guinea pig anti-rabbit IgG negative control antibody ABIN101961) to the respective tube, corresponding to a 1:100 dilution.
- Rotate the microcentrifuge tubes ON at 4 °C.
- Spin down the liquid and place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Resuspend with 1 mL Digitonin Wash Buffer and mix by inversion. If clumping occurs, gently remove the clumps with a 1 ml pipette tip.
- Repeat once for a total of two washes.
- pA-MNase Binding
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Vortex the sample at low speed and add 150 μL CUTANA pAG-MNase 0.5X (antibodies-online ABIN6950951, 1:40 dilution of a 20X stock in Digitonin Wash Buffer) per sample, gently resuspending the beads by pipetting.
- Rotate the microcentrifuge tubes for 1 h at 4 °C.
- Spin down the liquid and place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Resuspend with 1 mL Digitonin Wash Buffer and mix by inversion. If clumping occurs, gently remove the clumps with a 1 mL pipette tip.
- Repeat once for a total of two washes.
- MNase digestion and release of pA-MNase-antibody-chromatin complexes
- Spin down the liquid from the lid with a quick pulse in a table-top centrifuge.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Place each tube at a low angle on the vortex mixer set to a low speed and add 100 μL Digitonin Wash buffer per sample along the side of the tube.
- Place tubes in a heat block, kept on ice, and allow to chill.
- Add 2 μL 0.1 M CaCl2 to each sample.
- Incubate tubes at 0 °C for 15 min.
- Add 100 μL 2X STOP buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.05% (wt/vol) Digitonin, 100 μg/mL RNAse A, 50 μg/mL Glycogen).
- Incubate tubes at 37 °C for 30 min.
- Place the tubes on a magnet stand until the fluid is clear.
- Transfer the supernatant containing the pAG-MNase-bound digested chromatin fragments to fresh 1.5 mL microcentrifuge tubes.
- DNA extraction
- Add 2 µL 10% SDS to a final concentration of 0.1% and 2.5 µL Proteinase K (20 mg/mL) to a final concentration of 0.25 mg/mL to each supernatant.
- Gently vortex tubes at a low speed of approximately 1,100 rpm.
- Incubate tubes at 50 °C for 1 h.
- Add 200 µL PCI to tube.
- Vortex tubes thoroughly at high speed until the liquid appears milky.
- Centrifuge tubes in a tabletop centrifuge at 16,000 x g at RT for 5 min.
- Carefully transfer to upper aqueous phase to a fresh 1.5 mL microcentrifuge tube containing 2 µL glycogen (diluted 1:10 to 2 mg/mL from the 20 mg/mL stock solution).
- Add 20 µL 3 M NaOAc pH 5.2.
- Add 400 µL 100% ethanol.
- Place tubes for at -20 °C ON.
- Centrifuge tubes in a tabletop centrifuge at 16,000 x g at 4 °C for 5min.
- Remove the liquid carefully with a pipette.
- Wash pellet with 1ml 70% ethanol.
- Centrifuge tubes in a tabletop centrifuge at 16,000 x g at 4 °C for 1 min.
- Remove the liquid carefully with a pipette.
- Air-dry the pellet, then dissolve in 30 µL 1 mM Tris-HCl, 0.1 mM EDTA.
- Library preparation and sequencing
- Prepare Libraries using KAPA HyperPrep Kit using KAPA Dual-Indexed adapters according to protocol.
- Sequence samples on an Illumina NextSeq 500 sequencer, using a NextSeq 500/550 High Output Kit v2.5 (75 Cycles), 36bp PE.
- Peak calling
- Trim reads using bbTools bbduk to remove adapters, artifacts and repeat sequences.
- Aligned reads were mapped to the GRCh38 (hg38) human genome using bowtie2 with options --local --very-sensitive- local --no-unal --no-mixed --no-discordant - X 400.
- Convert SAM files to BAM files and remove duplicates using SAMtools was used to convert SAM files to BAM files. Produce Bedgraph files with BEDtools genomecov.
- Call peaks using SEACR against the IgG negative control with the options norm and relaxed.
- Experimental Notes
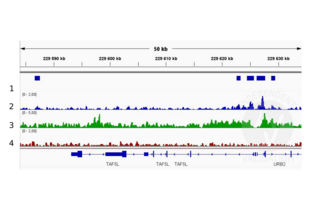
The CUT&RUN alignment track was compared to a reference alignment track of ChIP-seq for H3K4me in HEK293 cells obtained from ENCODE (PMID 26527727), experiment ENCSR000FCG, track ENCFF274LAP.
Validation #104228 (Cleavage Under Targets and Release Using Nuclease)![Successfully validated 'Independent Validation' Badge]()
![Successfully validated 'Independent Validation' Badge]() Validation Images
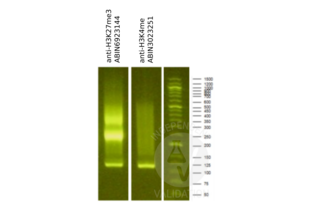
Validation Images![Library profiles comparing fragment size distributions on an E-Gel EX 2% agarose gel (Thermo Fisher). Fragments obtained from CUT&RUN using an anti-H3K27me3 CUT&RUN Positive Control antibody (ABIN6923144) and anti-H3K4me (ABIN3023251) after library preparation, compared to the E-Gel Sizing DNA Ladder (Thermo Fisher).]() Library profiles comparing fragment size distributions on an E-Gel EX 2% agarose gel (Thermo Fisher). Fragments obtained from CUT&RUN using an anti-H3K27me3 CUT&RUN Positive Control antibody (ABIN6923144) and anti-H3K4me (ABIN3023251) after library preparation, compared to the E-Gel Sizing DNA Ladder (Thermo Fisher).
Library profiles comparing fragment size distributions on an E-Gel EX 2% agarose gel (Thermo Fisher). Fragments obtained from CUT&RUN using an anti-H3K27me3 CUT&RUN Positive Control antibody (ABIN6923144) and anti-H3K4me (ABIN3023251) after library preparation, compared to the E-Gel Sizing DNA Ladder (Thermo Fisher).
![Alignment tracks from CUT&RUN targeting H3K4me in HEK293T cells. 1. Peaks called by SEACR from CUT&RUN data using anti-H3K4me antibody ABIN3023251. 2. Alignment track for CUT&RUN reads obtained using anti-H3K4me antibody ABIN3023251 in HEK293T cells. CUT&RUN reads are normalized to sequencing depth per million reads. 3. Alignment track of ChIP-seq for H3K4me in HEK293 cells obtained from ENCODE, experiment ENCSR000FCG, track ENCFF274LAP. Coverage is shown as fold change over control. 4. Alignment track for CUT&RUN negative control normalized to sequencing depth per million reads.]() Alignment tracks from CUT&RUN targeting H3K4me in HEK293T cells. 1. Peaks called by SEACR from CUT&RUN data using anti-H3K4me antibody ABIN3023251. 2. Alignment track for CUT&RUN reads obtained using anti-H3K4me antibody ABIN3023251 in HEK293T cells. CUT&RUN reads are normalized to sequencing depth per million reads. 3. Alignment track of ChIP-seq for H3K4me in HEK293 cells obtained from ENCODE, experiment ENCSR000FCG, track ENCFF274LAP. Coverage is shown as fold change over control. 4. Alignment track for CUT&RUN negative control normalized to sequencing depth per million reads.
Full Methods
Alignment tracks from CUT&RUN targeting H3K4me in HEK293T cells. 1. Peaks called by SEACR from CUT&RUN data using anti-H3K4me antibody ABIN3023251. 2. Alignment track for CUT&RUN reads obtained using anti-H3K4me antibody ABIN3023251 in HEK293T cells. CUT&RUN reads are normalized to sequencing depth per million reads. 3. Alignment track of ChIP-seq for H3K4me in HEK293 cells obtained from ENCODE, experiment ENCSR000FCG, track ENCFF274LAP. Coverage is shown as fold change over control. 4. Alignment track for CUT&RUN negative control normalized to sequencing depth per million reads.
Full Methods -
- Format
- Liquid
- Buffer
- PBS with 0.02 % sodium azide,50 % glycerol, pH 7.3.
- Preservative
- Sodium azide
- Precaution of Use
- This product contains Sodium azide: a POISONOUS AND HAZARDOUS SUBSTANCE which should be handled by trained staff only.
- Handling Advice
- Avoid freeze / thaw cycles
- Storage
- -20 °C
- Storage Comment
- Store at -20°C. Avoid freeze / thaw cycles.
-
-
: "A new cut&run low volume-urea (LoV-U) protocol optimized for transcriptional co-factors uncovers Wnt/b-catenin tissue-specific genomic targets." in: Development (Cambridge, England), (2022) (PubMed).
: "Functional characterization of rice CW-domain containing zinc finger proteins involved in histone recognition." in: Plant science : an international journal of experimental plant biology, Vol. 263, pp. 168-176, (2018) (PubMed).
: "Celastrol attenuates incision-induced inflammation and pain associated with inhibition of the NF-κB signalling pathway via SARM." in: Life sciences, Vol. 205, pp. 136-144, (2018) (PubMed).
: "Iron chelation inhibits cancer cell growth and modulates global histone methylation status in colorectal cancer." in: Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine, Vol. 31, Issue 5, pp. 797-805, (2018) (PubMed).
-
: "A new cut&run low volume-urea (LoV-U) protocol optimized for transcriptional co-factors uncovers Wnt/b-catenin tissue-specific genomic targets." in: Development (Cambridge, England), (2022) (PubMed).
-
- Target
- Histone 3 (H3)
- Alternative Name
- Histone H3 (H3 Products)
- Background
- Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H3 family. Transcripts from this gene lack polyA tails, instead, they contain a palindromic termination element. This gene is located separately from the other H3 genes that are in the histone gene cluster on chromosome 6p22-p21.3.,H3.4,H3/g,H3FT,H3t,HIST3H3,Histone H3,HIST1H3A,Signal Transduction,MAPK-Erk Signaling Pathway,MAPK-P38 Signaling Pathway,Epigenetics & Nuclear Signaling,Epigenetic Modifications,Methylation,Histone H3
- Molecular Weight
- 15 kDa
- Gene ID
- 8290
- UniProt
- Q16695
-

 (4 references)
(4 references) (1 validation)
(1 validation)



